
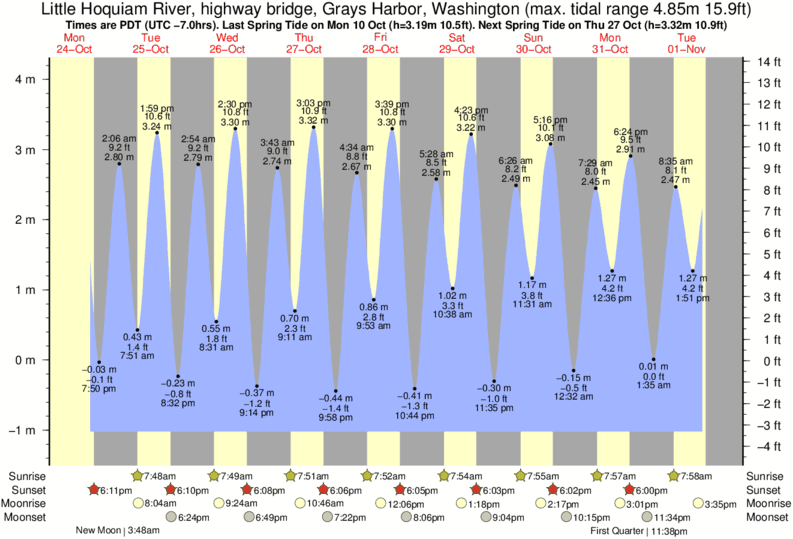
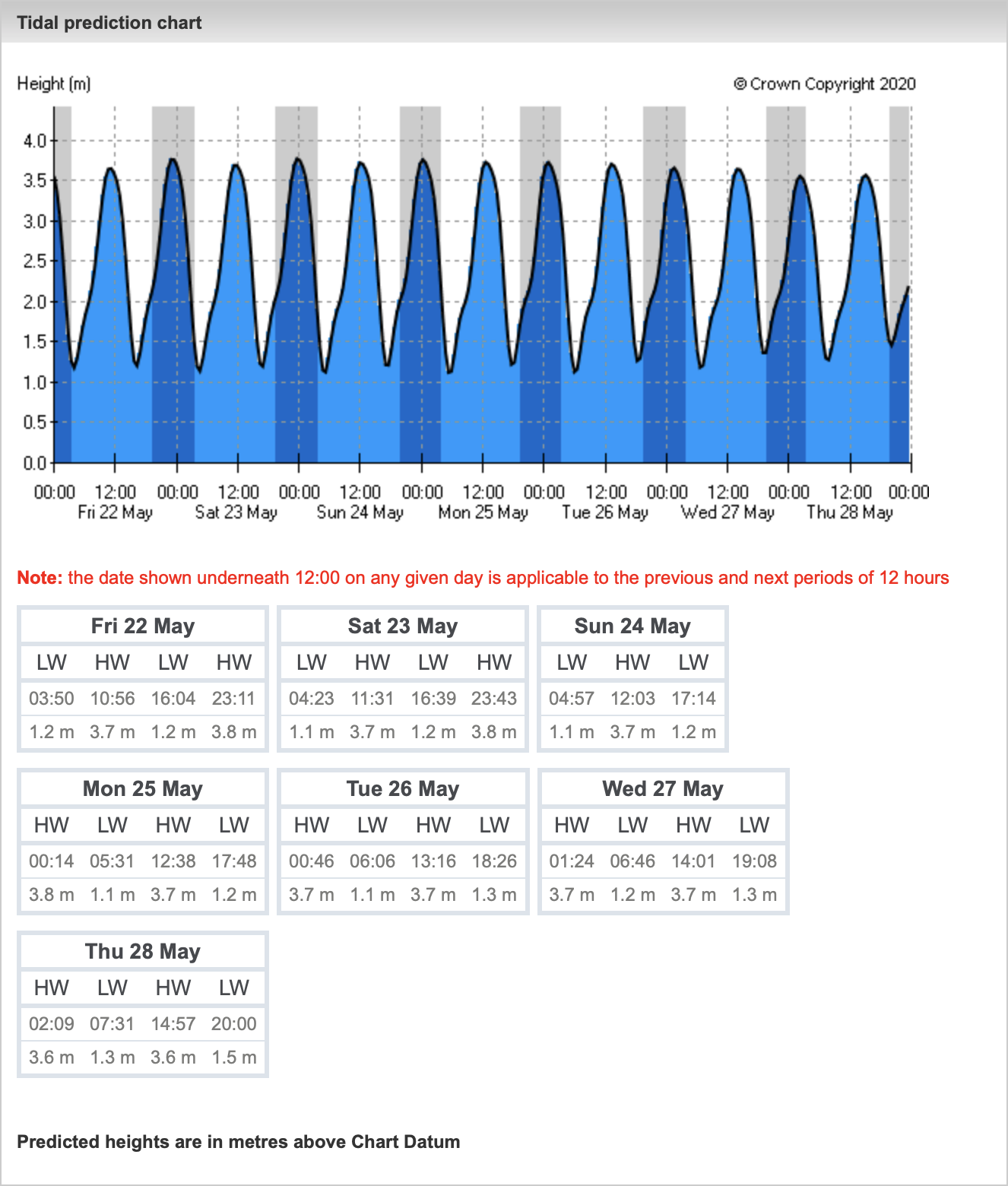
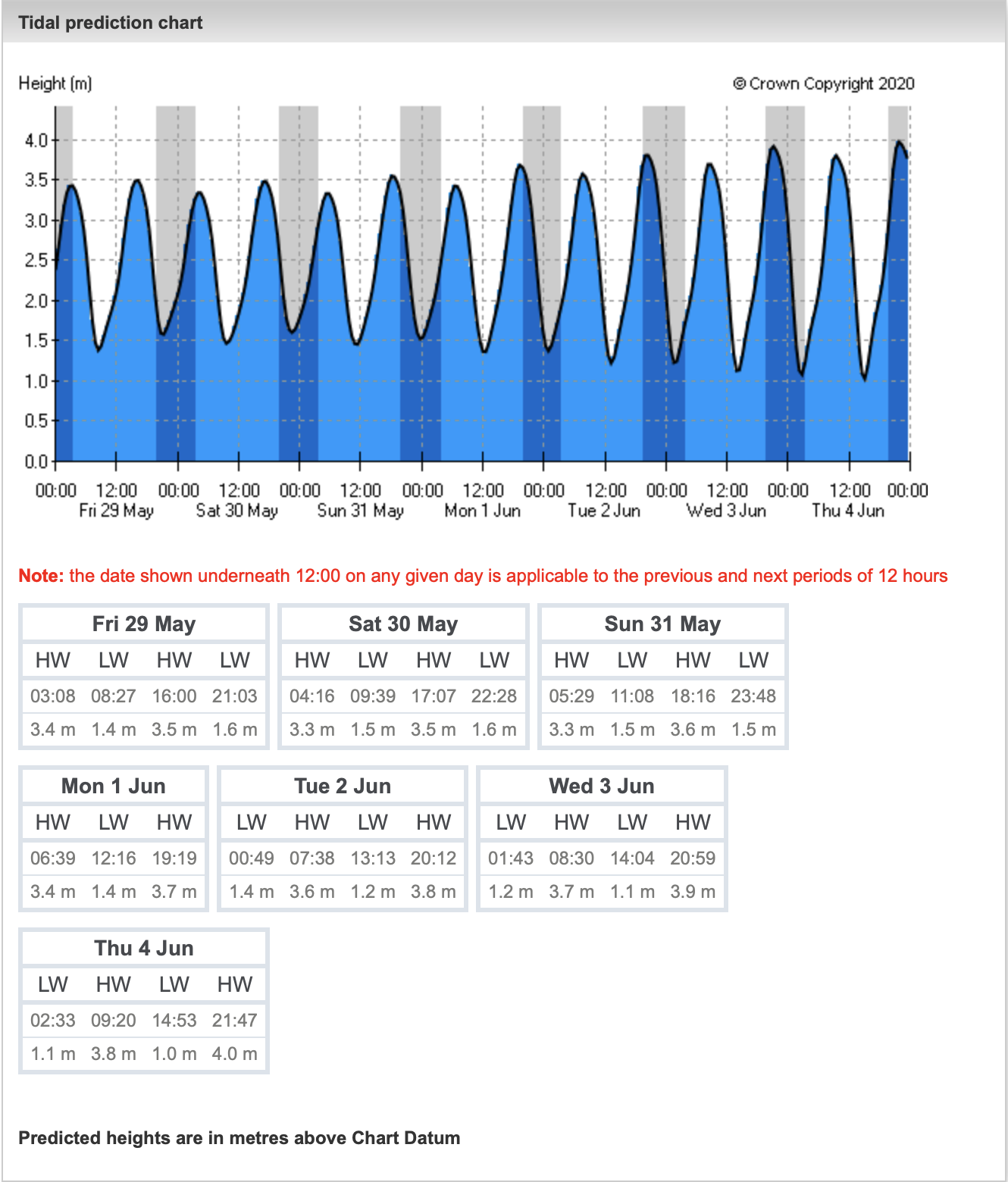
INPP5E, a phosphoinositide 5-phosphatase, localizes on the ciliary membrane via its C-terminal prenyl moiety, and maintains the distinct ciliary phosphoinositide composition.
Our findings reveal key players controlling early steps in aperture domain formation, identify residues important for their function, and open new avenues for investigating how diversity of aperture patterns in nature is achieved. We also show that a third ELMOD family member, ELMOD_E, can interfere with MCR and ELMOD_A activities, changing aperture morphology and producing new aperture patterns. We show that two members of this family, MACARON (MCR) and ELMOD_A, act upstream of the previously discovered aperture proteins and that their expression levels influence the number of aperture domains that form on the surface of developing pollen grains. Here, we demonstrate that patterns of aperture domains in Arabidopsis are controlled by the members of the ancient ELMOD protein family, which, although important in animals, has not been studied in plants. However, how these aperture domains are selected is unknown. During pollen development, certain plasma membrane domains attract specific proteins and lipids and become protected from exine deposition, developing into apertures. In each species, aperture number, position, and morphology are typically fixed across species they vary widely. Pollen apertures, the characteristic gaps in pollen wall exine, have emerged as a model for studying the formation of distinct plasma membrane domains. Thus, we believe that ELMOD1 and ELMOD3 perform multiple functions in cells, most prominently linked to ciliary biology and Golgi-ciliary traffic, and likely acting from more than one cellular location. These phenotypes are reversed upon expression of activating mutants of either AR元 or ARL16, linking their roles to ELMOD1/3 actions.
Deletion of either Elmod1 or Elmod3 results in the decreased ability of cells to form primary cilia, loss of a subset of proteins from cilia, and accumulation of some ciliary proteins at the Golgi, predicted to result from compromised traffic from the Golgi to cilia. We found strong similarities in phenotypes resulting from deletion of either Elmod1 or Elmod3 and marked differences from those arising in Elmod2 deletion lines. Here, using similar strategies with the paralogs ELMOD1 and ELMOD3, we identify novel functions and locations of these cell regulators and compare them to those of ELMOD2, allowing determination of functional redundancy among the family members. We described functions of ELMOD2 in immortalized mouse embryonic fibroblasts (MEFs) in the regulation of cell division, microtubules, ciliogenesis, and mitochondrial fusion. ELMODs are ubiquitously expressed in mammalian tissues, highly conserved across eukaryotes, and ancient in origin, being present in the last eukaryotic common ancestor. ELMODs are a family of three mammalian paralogs that display GTPase activating protein (GAP) activity towards a uniquely broad array of ADP-ribosylation factor (ARF) family GTPases that includes ARF-like (ARL) proteins.


 0 kommentar(er)
0 kommentar(er)
